
The CMD2020GEFES is a large international conference covering all aspects of condensed matter physics. CMD2020GEFES is an Europhysics Conference, as recognized by the Action Committee on Conferences of the European Physical Society.
The 28th series of the CMD2020GEFES was originaly planned to take place in Madrid between the 31st and 4th of August 2020, but due to the current circumstances the event is going to be held online. The full scientific program can be found in the conference website.
Quasar will join the conference with the presence of Jaime Carracedo-Cosme. Below we provide the sumitted abstract.
Join the event between the 31st August-4th September!
CMD2020GEFES ABSTRACT
Molecular identification based on AFM with CO tips and Deep Learning
Jaime Carracedo-Cosme1,2, Carlos Romero-Muñiz 2,3, Pablo Pou2,4, Rubén Pérez2,4
1 Quasar Science Resources SL, Camino de las Ceudas 2, E-28232 Las Rozas de Madrid, Spain
2 Departamento de Física Teórica de la Materia Condensada, Universidad Autónoma de Madrid, E-28049 Madrid
3 Departamento de Sistemas Físicos, Químicos y Naturales, Universidad Pablo de Olavide, E-41013 Sevilla, Spain.
4 Condensed Matter Physics Center (IFIMAC), Univ. Autónoma de Madrid, E-28049 Madrid, Spain
jcarracedo@quasarsr.com
Non-contact atomic force microscopy (NC-AFM) with CO-functionalized metal tips images the internal structure of molecules with unprecedented resolution, resolving intermolecular features and determining bond orders in aromatic compounds [1]. Its capability to address individual molecules has paved the way for the identification of natural compounds (where conventional methods failed) and of the intermediates and final products generated in on-surface reactions, and to resolve more than a hundred different types of molecules in complex molecular mixtures such as asphaltenes [1].
Here, we present our strategy to transform these potentialities into a robust characterization technology, combining high-resolution imaging and machine learning in order to achieve molecular identification. We focus on a large set of quasi-planar molecules that spans all the relevant structural and compositional moieties. We describe how, from each of these molecules, we build the dataset of 2D theoretical images needed for training and validation of our deep learning model, striking the right balance to incorporate enough variation and to prevent overfitting. The simulations of AFM images are based on a recent evolution of the probe-particle (PP) model [2], that evaluates the Pauli repulsion potential as an overlap of the electron densities of tip and sample (following our proposal in ref. [3] with exponent α=1), and represents the molecule by a quadrupole model to calculate the interaction with the sample electrostatic potential. The charge densities and the electrostatic potential are obtained from independent Density Functional Theory (DFT) calculations.
Next, we discuss the performance of several deep learning models to identify theoretical images, and analyze them in terms of the transfer of relevant information across the different layers in the model (Figure 1). Finally, we apply the model to the identification of experimental images. Here, we focus on understanding how elemental sensitivity can be boosted by identifying features in the images that reflect the highly anisotropic spatial decay of the molecular charge density and discuss the need for AFM image simulation methods that, while still fast, properly capture these effects [3].
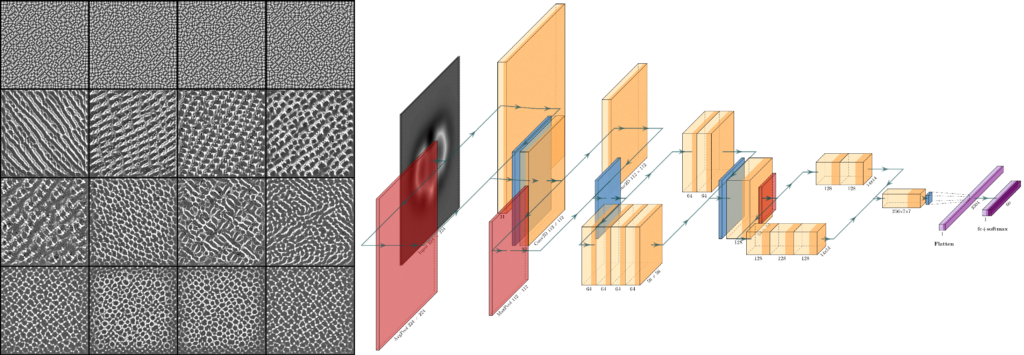
References
- L. Gross, et al., Angew. Chem.Int. Ed. 57, 3888 (2018).
- P. Hapala, et al., Phys. Rev. B 90, 085421 (2014).
- M. Ellner, et al., ACS Nano 13, 786 (2019).
VIDEO PRESENTATION
Non-contact atomic force microscopy (NC-AFM) consists in a complex process based on a simple idea; if we approach a cantilever finished in a tiny tip to one molecule, the tip will experiment attractive and repulsive forces. If we measure these forces and plot them, we obtain an image (see image below with Subfig B).
The CO-functionalized metal tip procedure consists in performing the previous calculation but using a CO (carbon+oxygen) molecule attached to the tip. The difference with previous results is reflected in Subfigs C and D in the image below. The main reason is that the tip finished with a CO is more flexible and can measure AFM images in greater detail. This is what is meant by “resolving intermolecular features”. Despite having improved these results with the CO-tip, it is not possible yet to identify what molecule was used to obtain the AFM image (if one did not know that image C was obtained from molecule A, it would be difficult to deduce this information from the AFM imageitself as, for example, one could replace a CH with a nitrogen and the AFM results would be similar). Our idea is to develop an automatic method (using deep learning techniques, a type of artificial intelligence) to identify the molecule seen in Subfig A, used to obtain the AFM image in Subfig C.

Our main objective in this work has been to see if a deep learning model is capable of differentiating molecules that differ in a single atom, ring distribution or number of ring vertices. For this purpose, we have selected a set of quasi-planar molecules with different atoms and rings distribution. Here, the term quasi-planar is important because forces are meassured by approaching the tip to the molecule so all the atoms mut be at the same height.
Deep learning needs large amounts of data. it is difficult to obtain enough experimental AFM data to adjust (train) our model. The solution that we proposed is to perform theoretical AFM simulations to generate the dataset. We have improved a simulation model (Probe Particle Model) by adding more detailed calculations from a model developed by us. Different model parameters are varied to obtain sets of different images (tip height, amplitude range, tilting stiffness) and we slightly rotate the position of the molecules to obtain different images.
To classify the images, first we have applied well-established deep learning models (VGG16 and MobileNetV2). We develop a suitable model to perform the AFM image classification.

