Quasar Science Resources collaborates with the Scanning Probe Microscopy Theory & Nanomechanics (SPMTH) research Group (Department of Theoretical Condensed Matter Physics at the Universidad Autonoma de Madrid (Autonomous University of Madrid, UAM) in the development of an efficient method to identify the structure and chemical composition of individual molecules based on the application of machine learning tools to Atomic Force Microscope (AFM) images. This project is supported by the Industrial Doctorate Program of the Comunidad de Madrid, that fosters the collaboration of Universities and Private Enterprises in the training of PhD students.

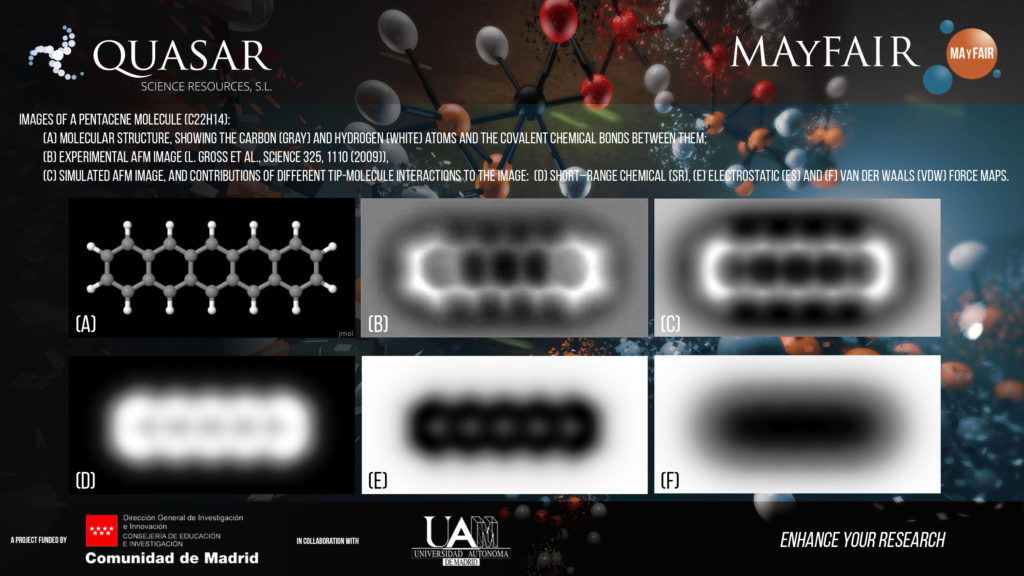
AFM senses the strength of the interaction between a sharp tip and a sample surface to build an image with nanometer resolution. AFM is one of the key tools that have supported the development of nanoscience and nanotechnology. Recent technical advances (operation in the frequency modulation (FM) mode, and the use of highly inert tips) have extended the AFM imaging capabilities to the atomic scale. It is now possible to resolve the location of individual atoms and the chemical bonds in AFM images of a molecule absorbed on an insulating surface (Figure 1b). The bright areas in the experimental image closely resemble the structure of the pentacene molecule (see the ball–and-stick model in Figure 1a), the first molecule imaged with FM-AFM. This technique is already revolutionizing the emerging field of on–surface chemistry and has the potential to become a key tool for molecular identification where standard techniques like magnetic resonance (NMR) fail.
The aim of this project is precisely to lead this breakthrough, providing a reliable, robust, and efficient method to identify the structure and chemical composition of individual molecules from information that can be gathered with the AFM. Our approach to accomplish this ambitious goal combines two key elements: (i) the use and further development of an efficient simulation tool for theoretical AFM images (Figure 1c), based on the expertise of the SPMTH group. This tool will help us to understand the mechanisms that controlled the contrast in AFM images, and, in particular, the role of the different contributions to the total tip-molecule interaction (Figures 1d, 1e, 1f), and (ii) the development of advanced computational techniques to store, classify and process experimental data using machine-learning tools in order to enhance the predictive power of our identification method.